A human being consists of approximately 60 trillion (60x10^12) cells. At
each instant, in each of these 60 trillion cells, the genome, a ribbon of 2
billion characters, is decoded to produce the proteins needed for the survival
of the organism. This genome contains the ensemble of the genetic inheritance
of the individual and, at the same time, the instructions for both the construction
and the operation of the organism. The parallel execution of 60 trillion genomes
in as many cells occurs ceaselessly from the conception to the death of the individual.
Faults are rare and, in the majority of cases, successfully detected and repaired.
This process is remarkable for its complexity and its precision. Moreover, it
relies on completely discrete information: the structure of DNA (the chemical
substrate of the genome) is a sequence of four bases, usually designated with
the letters A (adenine), C (cytosine), G (guanine), and T (thymine).
Our Embryonics project (for embryonic electronics ),
situated on the ontogenetic axis of our POE
model , is inspired by the basic processes of molecular biology and
by the embryonic development of living beings. By adopting certain features
of cellular organization, and by transposing them to the two-dimensional world
of integrated circuits on silicon, we will show that properties unique to the
living world, such as self-replication and self-repair, can also be applied
to artificial objects (integrated circuits).
Objectives and Strategy
Our final objective is the development of very large scale integrated (VLSI)
circuits capable of self-repair and self-replication .
Self-repair allows partial reconstruction in case of a minor fault, while self-replication
allows complete reconstruction of the original device in case of a major fault.
These two properties are particularly desirable for complex artificial systems
in situations which require improved reliability, such as :
- Applications which require very high levels of reliability, such as avionics
or medical electronics.
- Applications designed for hostile environments, such as space, where
the increased radiation levels reduce the reliability of components.
- Applications which exploit the latest technological advances, and notably
the drastic device shrinking, low power supply levels, and increasing operating
speeds, which accompany the technological evolution to deeper submicron
levels and significantly reduce the noise margins and increase the soft-error
rates.
- Applications that will need to be executed flawlessly on highly-flawed
substrates, such as the future nano-electronic devices, whose characteristics
will be very close to those of living organisms.
These emerging needs require the development of a new design paradigm that supports
efficient online testing and self-repair solutions. Drawing inspiration
from the architecture of living beings, we wish to show how to implement online
testing, self-repair, and self-replication using both hardware and software redundancy.
Approach
Within the Embryonics project, we have been studying the application of biological
ontogenesis to the design of digital hardware for several years. Among what
we feel are our main contributions to the field is a self-contained representation
of a possible mapping between the world of multi-cellular organisms in biology
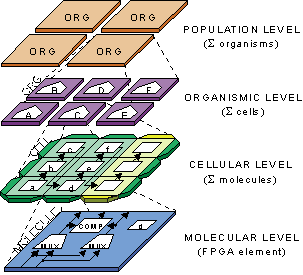
and the world of digital hardware systems, based on 4 levels of complexity,
ranging from the population of organisms to the molecule.
Within this mapping, we define an artificial organism as
a parallel array of cells, where each cell is a simple processor
that contains the description of the operation of every
other cell in the organism in the form of a program (the
genome). For a given application, all
cells are structurally identical and contain the same program
(and can thus be seen as stem cells), but different parts
of the program and of the structure are activated depending
on the cell's position within the array, implementing cellular differentiation.
Moreover, our cells are application-specific, that is, are structurally and
fuctionally adapted to the application to be executed, again in keeping with
our biological inspiration (a skin cell
is physically different from a liver cell). To achieve this property, we redefined
our cells as reconfigurable processing elements, realized by custom programmable
logic circuits (FPGA) that fulfil the role of molecules in
our system. Dedicated hardware within our FPGA implements special features, such
as growth, adaptation, and self-repair, that are both highly unusual
in computing devices and potentially useful in view of the latest technological
advances.
|
|
Text from https://en.wikipedia.org/wiki/Embryonics_Project CC-By-SA 3.0
Contacts:
Prof. Daniel MANGE
Prof. Gianluca TEMPESTI
Key publications:
