
Darwin Initiative for the Survival of
Species
(UK Government - DETR)
 |
Darwin Initiative for the Survival of
Species
|
Collaborating Institutions: Earth Biosphere Institute and School of Biology,
University of Leeds, UK; Department of Biology, University of York, UK, Institute
of Tropical Biology & Conservation, University of Malaysia Sabah; Forest
Research Centre, Sepilok, Sabah
Personnel: Project leader - Dr Keith Hamer. Fieldwork manager
- Dr Jane Hill.
Molecular genetics manager - Dr Jeremy Searle. Malaysia co-ordinator
- Prof Maryati Mohamed.
Research advisor - Dr Chey vun Khen.
Darwin Research Fellows - Suzan Benedick, Nazirah Mustaffa
Tropical conservationists face three major challenges in identifying priorities for conservation in the face of limited funding resources and the continuing loss of biodiversity: (1) tropical communities contain many species, especially invertebrates, that have not bee formally identified and whose taxonomy is only poorly resolved; (2) limited resources need to be targeted towards those species with the highest conservation value, but many tropical countries lack the molecular genetic skills required to identify those species that have the greatest phylogenetic uniqueness and so contribute most to biodiversity; (3) isolation of populations within forest fragments can limit the dispersal of individuals, so reducing genetic diversity within populations and increasing the likelihood of local extinctions. Effective conservation in fragmented landscapes requires estimates of gene flow patterns within and among habitat fragments, in order to minimize the isolation of populations and consequent loss of biodiversity. Yet the molecular genetic tools for estimating gene flow patterns are lacking in many tropical countries. This is a matter of urgent concern because in the near future, most remaining tropical forest will occur as fragments scattered among agriculture and urban development.

The Malaysian State of Sabah lies at the northern tip of Borneo
The State of Sabah (Borneo) is the poorest financially in Malaysia and most of its income is generated through conversion of rainforest into oil palm plantation and other forms of silviculture. This increasingly leaves patches of rainforest interspersed among oil palm and other plantations. These rainforest patches may contribute significantly to the conservation of rainforest biodiversity and some have been gazetted as Forest Reserves to protect them from further disturbance. However resources for protection are limited and forest managers lack the means of establishing priorities for patches based on key criteria (species diversity, representation of species of high conservation value and importance within a network of local populations). This lack of vital information also excludes forest managers from making informed recommendations as to the size and placement of forest patches to be preserved in future agricultural developments.
The main purpose of the project is, in consultation with Sabahan forestry researchers
and managers, to use molecular genetic tools to further the development of strategies
that balance conservation of rainforest biodiversity with agricultural development
to meet local community requirements. To these ends, the project aims to attain
the following major objectives:
(i) to train two Sabahan graduate biologists (Darwin Research Fellows) in molecular
techniques for resolving taxonomy and quantifying genetic diversity, and in
ecological techniques for sampling and monitoring biodiversity in forest patches;
(ii) to produce a comprehensive quantitative inventory of butterfly species
in different sized forest fragments;
(iii) to establish a number of semi-permanant study plots in forest fragments
for long-term monitoring of butterfly biodiversity;
(iv) to evaluate the relationship between forest fragmentation (size and isolation)
and the diversity and conservation value of forest-dwelling butterfly species;
(v) to use molecular genetic techniques to resolve the taxonomy of problematic
butterfly species and to attach conservation values to species on the basis
of phylogenetic (evolutionary) distinctiveness;
(vi) to estimate gene flow and genetic population structure of selected butterfly
species.
The study focuses on 10 lowland rainforest sites, varying in size from 1.2km2 to 10,000km2.
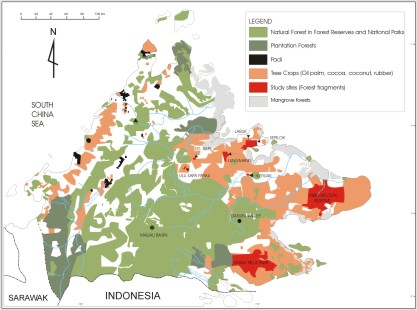
Map to showing the study sites. Forest fragments are shown
in red.
The two control sites are Danum Valley and Maliau Basin.
At each site, we are using three different techniques to sample a variety of
species:
(1) Fruit-feeding butterflies are sampled using traps baited with fruit;
(2) Carrion-feeding butterflies are sampled using traps baited with rotting
fish and prawn paste;
(3) Family Lycaenidae (blues) are sampled by hand-netting along transects at
each site.
|
|
|
We are also sampling butterflies in oil palm plantation, using all three sampling
methods above, to provide information on the ability of species to disperse
between forest fragments embedded in a matrix of plantations.
In most cases, captured butterflies are identified in the field, marked with
a unique number or a coloured dot (depending on body size) and released. All
individuals of new species, not previously sampled, are collected and preserved.
All these specimens are housed in the permanent insect collections at Universiti
Malaysia Sabah and the Forest Research Centre Sabah. In addition, we are collecting
a sample of individuals from selected species for molecular genetic analysis.
These include problematic species (genera Taenacia and Euthalia)
and all species in the genus Mycalesis: this is one of the most species-rich
genera of butterfly in Sabah and contains a mixture from endemic species (M.
kina, M. amoeba) to species with very broad geographical distributions (e.g.
M. mineus, occurring in both Oriental and Australasian regions), making
it an ideal model genus to study phylogenetic distinctiveness. In addition,
one species of Mycalesis (M. orseis) is confined to dense forest
habitats but occurs at all study sites, making it an excellent model species
to estimate gene flow and genetic structuring of populations. Material for molecular
genetic analysis is being transported to the UK, under licence, for extraction,
amplification and sequencing of mitochondrial DNA using standard techniques.

Mycalesis kina - a species endemic to Borneo
Click here for useful link to: